The synthesis of DNA fragments by the phosphoramidite triester method has the characteristics of efficient and rapid coupling and stable initial reactants. The phosphoramidite triester method is to immobilize DNA on a solid phase carrier to synthesize DNA strands. The synthesis direction is synthesized from the 3' end to the 5' end of the primer to be synthesized, and the adjacent nucleotides pass through 3'. → 5' phosphodiester bond linkage.
The first step is to react the protected group-protected nucleotide pre-attached to the solid phase carrier CPG with trichloroacetic acid to remove the 5'-hydroxyl protecting group DMT to obtain a free 5'-hydroxyl group.
In the third step, a capping reaction, there may be a very small number of 5'-hydroxy groups in the condensation reaction that do not participate in the reaction (less than 2%), and the reaction continues with acetic anhydride and 1-methylimidazole. Short fragments can be isolated upon purification.
In the fourth step, the phosphorous acid form is converted to a more stable phosphotriester by the action of the oxidant iodine.
After the above four steps, a deoxynucleotide is attached to the nucleotide of the solid phase carrier. The protective group DMT on its 5'-hydroxyl group is then removed with trichloroacetic acid and the above procedure is repeated until all of the bases required to be synthesized are attached. The color determination synthesis efficiency of the TCA processing stage can be observed during the synthesis.
The primers attached to the CPG were excised by high temperature treatment of ammonia water, and the primers were purified by OPC, PAGE, etc., and the finished primers were concentrated, desalted, and precipitated by C18. The precipitated primers were suspended in water, and the OD260 was quantified and dispensed according to the order requirements.
2. What are the methods of purifying the primers and how to choose them?
3. How is the OD number of the primer quantified?
A: The OD number of primer synthesis primers was determined by measuring the optical density of the solution using an ultraviolet spectrophotometer, a wavelength of 260 nm, a quartz cuvette, and an optical path of 1 cm. The optical density of the solution is preferably diluted to between 0.2 and 1.0. After the DNA dry powder was sufficiently shaken and dissolved with a certain volume of water, the OD value was measured by diluting with 1 ml of water. The OD value of the mother liquor needs to be converted according to the dilution factor.
4. What level of primers are needed?
A: The commonly used purification methods are desalting, BioRP / OPC purification, PAGE purification, and HPLC purification. Determine the purity level of the ordered primers based on the needs of the experiment.
application | Primer length requirement | Purity level requirement |
General PCR amplification | <45 base | BioRP / OPC |
General PCR amplification | >45 base | PAGE |
Diagnostic PCR amplification | < 40base | BioRP / OPC, PAGE |
DNA sequencing | Around 20base | BioRP / OPC |
Subcloning, point mutation, etc. | According to the experimental requirements | BioRP / OPC, PAGE, HPLC |
Gene construction (full gene synthesis) | According to the experimental requirements | PAGE |
Antisense nucleic acid | According to the experimental requirements | PAGE |
Modified primer | According to the experimental requirements | PAGE, HPLC |
A: The longer the primer, the greater the probability of a problem. Some companies have synthesized 120base primers with very low yields. Unless necessary, it is recommended that the length of the synthetic fragment should not exceed 80 mer. According to the current primer synthesis efficiency, the percentage of the full length (not necessarily correct) of the 80 mer crude product will not exceed 40%, and there will be a lot of loss in subsequent processing. Very low.
6. How many OD numbers do you need to synthesize ?
A: Determined according to the purpose of the experiment. General PCR amplification, 2 OD primers, can do 200-500 50ul standard PCR reactions. If you are doing gene splicing or annealing to make a connection, 1 OD is enough. But some researchers do a few PCRs, but they need 5-10 OD. Primers for whole-genome construction are relatively long, but some of our researchers also require high OD numbers. The longer the segment, the lower the final full-length yield and the greater the chance of error. Exceeding the requirement of OD number is actually a waste of social resources. It also reflects the lack of self-confidence of some researchers, especially novices. It always feels that it needs to be repeated many times to succeed.
7. How to check the purity of the primer?
A: The convenient way to do this in the lab is to use the PAGE method. Electrophoresis was carried out using a 16% polyacrylamide gel supplemented with 7 M urea. Take 0.2-0.5 OD primers, dissolve with urea saturated solution or add urea dry powder to the primer solution until saturation, heat denaturation before loading (95 ° C, 2 mins). The purpose of adding urea is to denature, and the second is to increase the specific gravity of the sample and to easily add the sample. Electrophoresis was carried out at a voltage of 600 V. After a certain period of time (about 2-3 hours), the stripping was performed. The strip type was detected under a UV lamp using a fluorescent TLC plate, and there was no miscellaneous band under the main strip, indicating that the purity was good. If conditions permit, it can also be stained with EB staining or silver staining.
8. How to calculate the concentration of the primer?
A: Primers are stable at high concentrations. The primers are generally formulated to be 10-50 pmol/ul. In general, it is recommended to set the concentration of the primer to 50 pmol/ul, and the volume of water (microliter) is calculated as follows: V (microliter) = OD number * (multiplication) 33 * (multiplication) * (multiplication) 20000 / (except) the molecular weight of the primer. The molecular weight of the primers can be obtained from the synthesis report. If you need to prepare Other concentrations, convert according to the above formula.
Note: 1 OD260 = 33 ug/ml.
9. How to calculate the Tm value of the primer ?
A: Primer design software can give Tm, which is related to the length of the primer, the base composition, and the ionic strength of the primer using buffer.
For primers with a length of 25 mer or less, the Tm is calculated as: Tm = 4 ° C (G + C) + 2 ° C (A + T)
For longer oligonucleotides, the Tm is calculated as:
Tm = 81.5 + 16.6 x Log10[Na+] + 0.41 (%GC) – 600/size
In the formula, Size = the length of the primer.
Definition of Tm: Tm = Temperature at which 50% of a given oligonucleotide is hybridized to its complementary strand. In the absence of destabilizing agents, like formamide or urea, Tm will depend on 3 major parameters: The sequence: a GC-rich sequence The salt concentration: high oligonucleotide concentrations favor hybrid formation, which results in a higher melting temperature. The salt concentration: high ionic strength results in a higher Tm as cations stabilize the DNA duplexes.
10. How is the molecular weight of the primer (including modification) determined ?
A: The Molecular Weight of the unmodified primer is clearly indicated on the report provided with the primer. If it is necessary to estimate the molecular weight of one primer, the average molecular weight per base is 324.5, and the molecular weight of the primer = the number of bases x the average molecular weight of the base. Or calculate MW= (NA * WA) + (NC * WC) + (NG * WG) + (NT * WT) + (Nmod * Wmod) + (Nx * Wx) + ( Ni* Wi) +16 * Ns– 62.
NA, NG, NC, NT, Ni are the number of bases A or G or C or T or I in the primer, respectively, WA, WC, WG, W, Wi are the base A or G or C or T in the primer, respectively. The molecular weight of I, Nmod, Wmod are the number and molecular weight of the modifying groups, respectively.
The molecular weight of the mixed base is the sum of the molecular weights of the mixed bases divided by the number of mixing, for example, the molecular weight of the mixed G+A is (313.21 + 329.21) / 2 = 321.21. Ns is the number of thio groups, and the thio group increases the molecular weight by 16 at each position.
Conventional base molecular weight:
Base | Molecular Weight |
A | 313.21 |
C | 289.18 |
G | 329.21 |
T | 304.19 |
I | 314.2 |
U | 290.17 |
Modified group | Molecular weight | Modified group | Molecular weight | |
5'-Biotin | 405.45 | 3'-TAMARA | 623.60 | |
5'-(6 FAM) | 537.46 | 3'-Dabsyl | 498.49 | |
5'-HEX | 744.13 | 3'-(6 FAM) | 569.46 | |
5'-TET | 675.24 | 3'-Amino Modifier C3 | 153.07 | |
5'-Cy5 | 533.63 | 3'-Amino Modifier C7 | 209.18 | |
5'-Cy3 | 507.59 | 3'-Thiol Modifier C3 | 154.12 |
Answer: The material after drying is very loose. It is best to centrifuge before opening the lid, or the tube is tapped vertically on the table to collect the primer powder to the bottom of the tube. Add deionized sterile water or 10 mM Tris pH 7.5 buffer according to the calculated volume, leave it at room temperature for a few minutes, shake to help dissolve, and centrifuge to collect the solution to the bottom of the tube. The water used to dissolve the primers generally does not use distilled water, because some distilled water has a relatively low pH (pH 4-5), and the primers are unstable under such conditions.
12. How to save primers?
A: After the primers are synthesized, they are spin-dried to form a sheet-like substance after a series of treatment and purification steps. Primers can be stored for a long time at room temperature before dissolution. The dissolved primers -20 degrees can be stored for a long time. If the repeatability of the experiment is high, the number of synthesized ODs is large, and it is recommended to pack and avoid repeated freezing and thawing. Modification of fluorescent primers requires preservation in the dark.
13. The synthetic primers 5 'end phosphorylated whether there <br> A: synthetic primer 5' is a hydroxyl group, there is no phosphate group. If you need to use the polynucleotide kinase for 5' phosphorylation, or require the primer synthesis company to synthesize directly at the 5' or 3' end, additional charges are required.
14. What is the problem with the primer fragment being unable to attach to the carrier after annealing?
The ligation reaction requires the 5' phosphate group of the primer. If the synthesized primers need to be annealed directly to the corresponding vector, the primers need to be phosphorylated. If the phosphorylated product is not yet attached to the carrier, it is necessary to examine the enzymatic cleavage effect of the vector, and it is necessary to improve the conditions for primer annealing. SiRNA molecules have a special symmetrical structure, which is difficult to anneal, and the annealing temperature needs to be increased during annealing.
15. Sequencing found that there are mutations in the primers?
A: Sequencing found that there are mutations in the primer region, especially primers below 40 bases. The probability of occurrence is small, but it will definitely happen. Users can generally rest assured that the primer sequence is usually COPY directly to the synthesizer through the computer, and there are not many opportunities for base error. Nucleic acid synthesis companies generally have a set of controls to prevent base entry errors. There are many explanations for the reasons for this mutation, and there is no way to completely solve this problem. The principle of solid phase synthesis for primer synthesis is the same, the machines used are basically the same, and the main raw materials for synthesis are provided by a number of multinational companies. The problems encountered by each of the synthetic service providers are basically similar. No one can Super off.
Primer synthesis is a multi-step chemical reaction with a maximum synthesis efficiency of 99%, and by-products are unavoidable. The insertion mutation in the primer sequence is often a base repeat. It is generally believed that during the coupling process, part of the monomer that is being coupled is lost in DMT, causing the monomer to be connected again, so that a mutation inserted into the same base occurs. As for the deletion mutation, it is generally considered to be caused by the incomplete reaction of the capping reaction, and the caping reaction mainly blocks a very small number of 5'-hydroxy groups without participating in the reaction monomer. The closed primer will not continue to participate in the synthesis in the next round of coupling. For the base substitution mutation, the reason is generally believed that the base cannot be 100% deprotected, that is, the primer may contain a residual protecting group, and these regions of the primer are not well matched with the complementary strand when the amplified product is Subclones are transformed into E. coli and may be supplemented with unpaired bases by the bacterial repair system. Substitution mutations usually occur when G is converted to other bases. Base G can be converted to the enol isomer (dislocation) under certain conditions, 2,6 diaminopurine. DNA polymerase treats 2,6 diaminopurine as base A during DNA replication and amplification, and sequencing will reveal Base GA substitution. Depurination occurs more frequently in sputum-rich primers. If the depurinated primer is degraded during the post-primer deprotection phase, sequencing will reveal a deletion of base G or A.
During the synthesis of primers, the factors causing base insertion, deletion, and substitution mutations exist objectively. There are many suggestions and measures to reduce the frequency of occurrence, but these measures are still in the laboratory stage and have not yet been applied to large-scale production.
16. Why are long-chain primers having a high probability of error?
Answer: When the primers are synthesized, the efficiency of each step cannot reach 100%. The objective conditions for the factors of base insertion, deletion and substitution mutation are always present. The longer the primer chain, the higher the frequency of mutations. Researchers always hope that the synthesized primers will be foolproof, and this mood can be understood. However, as with PCR amplification, it is impossible to absolutely guarantee that there are no mutations in the amplified product, and primer synthesis cannot guarantee 100% correctness. It is important to know that the frequency of errors (non-human factors) in primer synthesis is higher than that produced by any high-fidelity high-temperature polymerase PCR amplification process. Do primer synthesis, long-chain primer synthesis, you have to prepare some of the primers may have mutations.
17. What if the mutation finds a mutation?
A: In this case, first contact with the nucleic acid synthesis manufacturer, the production staff will check the original record of production, mainly to check whether the synthetic sequence is consistent with the order, they will generally retain all the original data in the computer. In the case of confirming that the primer synthesis sequence is not mistyped, it is recommended to pick the clone and re-sequence and possibly find the correct clone. According to experience, 1-2 clones can be detected with primers below 40 bases; more than 40 primers, especially for full-splicing splicing, need to be tested. In general, the location of each clone mutation is different, and the prompt is always there, that is, how to find it. You can also ask the company to double the primers for free, but the coincident primers, like the first primers, may contain mutations that do not reduce your chances of encountering problems due to overlapping primers. In the process of gene splicing, if a small number of mutation points are found in a region, measure a few more, otherwise the primers will be overlapped.
18. Primers are purified by PAGE . Why are there base deletions or insertions?
A: Theoretically, analytical PAGE denaturing electrophoresis can distinguish the difference between one base and one base. However, the preparation of PAGE electrophoresis, the amount of loading is very large, the strips during electrophoresis are very wide, the overlap between the strip and the strip, the resolution has been reduced, and it is difficult to say that the difference is not cut when the target is removed after electrophoresis. A few base primers. There is a bad phenomenon in China. The amount of primers for PAGE purification, especially for long primers, is relatively high, which may result in a wider strip when cut. Recommendation: If you reduce the number of ODs, the primers may experience fewer problems.
19. What are the basic principles of TaqMan probe design?
A: The following principles are for your reference.
â—† The TaqMan probe position is as close as possible to the amplification primer (amplification product 50-150 bp), but it cannot overlap with the primer.
â—†The length is generally 18-40mer.
â—†GC content is controlled at around 40-80%.
â—† Avoid the appearance of consecutive identical bases, especially to avoid the appearance of GGGG or more G.
â—† Avoid using G at the 5' end of the primer.
â—† Use more bases C.
â—† Annealing temperature Tm is controlled at around 68-70C.
Useful fluorescent dye parameters
Name | Absorption wavelength | Emission wavelength | Colors |
6-FAM (6-carboxy-fluorescein) | 494nm | 518nm | Green |
TET (5-tetrachloro-fluorescein) | 521nm | 538nm | Orange |
HEX (5-hexachloro-fluorescein) | 535nm | 553nm | Pink |
TAMRA(tetramethyl-6-carboxyrhodamine) | 560nm | 582nm | Rose |
ROX (6-carboxy-x-rhodamine) | 587nm | 607nm | Red |
Cy3 (Indodicarbocyanine) | 552nm | 570nm | Red |
Cy5(Indodicarbocyanine) | 643nm | 667nm | Violet |
A: The following principles of primer design are for your reference.
â—† Primer length is generally 18-35mer.
â—†GC content is controlled at around 40-60%.
â—† Avoid enzyme cleavage sites or hairpin structures near the 3' end.
â—† If it is possible to avoid having more than 2 G or C in the last 5 bases of the 3' end.
â—† If it is possible to avoid the last base in the 3' end is A.
â—† Avoid the appearance of consecutive identical bases, especially to avoid the appearance of GGGG or more G.
â—† Annealing temperature Tm is controlled at about 58-60C.
â—† If you are designing a point mutation primer, the mutation point should be as close as possible to the middle of the primer.
21. Why is the primer's OD260/OD280 less than 1.5 ?
A: Primers should be all DNA, but why is the ratio of OD260/OD280 so low, how can there be protein contamination? Primer chemical synthesis, where is the opportunity to contaminate proteins? It should be noted that the ratio of OD260/OD280 cannot be used to measure the purity of the primer. The ratio of OD260/OD280 is too low due to the high content of C/T in the primer. The table below shows the ratio of OD260/OD280 of a 20mer homopolymer primer, clearly indicating that the ratio of OD260/OD280 is closely related to the base composition of the primer.
A260/280 ratios of Crude 20-mer Oligos of Differing Base Compositions
Base Composition | A260/280 |
5-AAAAAAAAAAAAAAAAAAAA-3 | 2.50 |
5-GGGGGGGGGGGGGGGGGGGG-3 | 1.85 |
5-CCCCCCCCCCCCCCCCCCCC-3 | 1.15 |
5-TTTTTTTTTTTTTTTTTTTT-3 | 1.14 |
5-AAAAAGGGGGTTTTTCCCCC-3 | 1.66 |
A: The amount of double-stranded DNA (such as plasmid DNA) can usually be judged by EB staining because EB can be chimeric into double-stranded DNA. However, the synthesized single-stranded DNA has different possibility of forming a secondary structure due to different base composition, and the degree of staining of EB is also different. For example, Oligo (dT) does not form a secondary structure, and EB staining effect is very poor. Therefore, do not use EB staining to quantify, but use UV spectrophotometer to detect. By the same token, photographs with EB staining are not suitable for all primers.
23. What are the consequences of primer impure?
A: Primer impureness may result in: 1) non-specific amplification; 2) inability to use enzymes pre-designed at the 5' end of the primer, especially primers without protecting the base; 3) for sequencing There is a double peak or a chaotic peak. Solution Resynthesis or repurification.
24. The primers that have been dissolved, why are they used normally, and it is not good to use them after a while?
A: If the pH of the dissolved primer is too low or contaminates the bacteria or nuclease, the primer will degrade. When used, the mixture is not fully thawed, and uneven liquid may cause inaccurate primer addition. It is recommended to dispense primers to avoid repeated freeze-thawing. It is recommended to use 10 mM Tris pH 7.5 buffer to dissolve the primers, as some distilled waters have a lower pH (pH 4-5) and the primers are not stable under these conditions. There is also a possibility that the primer has no problem, but the quality of the PCR material used, in particular the template, is not completely consistent with the previous use.
25. Is there a problem with the primers when PCR amplification is not available?
A: Basically not. Nowadays, various PCR amplification technologies have been developed, and various high-temperature polymerases are used to solve the problems of expansion and low amplification efficiency encountered in PCR amplification. For example, nested PCR is to amplify those gene fragments with very low copy number. Amplification of some repeat fragments, fragments with high GC content, must be amplified by special amplification methods.
Primers were not amplified, mainly in the following two cases (1) RT-PCR. Please note that many genes are difficult to amplify by conventional RT-PCR methods. The key to the success of RT-PCR is the RNA quality of the RT reaction and the content of the target gene in specific tissues and cells. (2) Amplification from the genome. In general, genes are single copies in the genome, and the genome needs to be strictly controlled as a template. The genomic DNA is too high, which affects the Mg and pH in the reaction system.
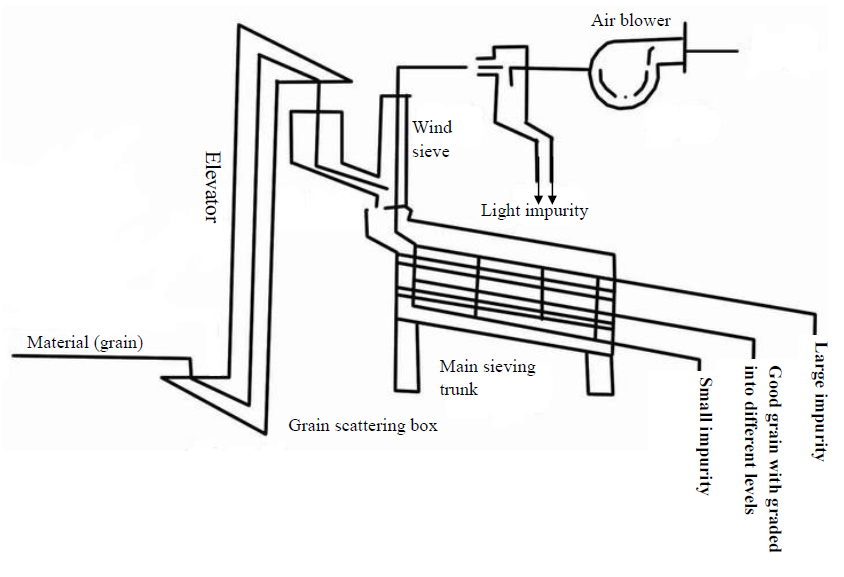
Seed Cleaner & Grader becomes basic and popular grain seed cleaning equipment in recent years. It helps farmers effectively clean grain harvested from ground. Seed cleaner & grader is used to remove oversize impurity, undersize impurity, light impurity from grain seed. Besides this, it also can grade cleaned seeds to different size ranks. From sesame, wheat, paddy, to soybean, kidney bean, groundnut, Seed Cleaner Cum Grader has wide application.
We have different Grain Cleaning Machine models. Capacity ranges from 3 ton per hour to 20 ton per hour. Each model has its particular feature. We will suggest client suitable model for particular seeds. Grain seed cleaner driving power is 3 phase electricity. We can customize grain cleaner base on different electricity specification for clients worldwide.
Grain cleaner has reliable running, convenient screen replacing and maintenance.
Seed
cleaning machine working procedure: Raw material is
fed from the Bucket Elevator, then elevator transfer raw material to the wind
sieve (air screen). In wind sieve, the straw, smashed pieces, dust and other
light impurity is sucked out by air blower. After air cleaning, raw material is
delivered to sieving trunk. Sieving trunk has several sieve layers. From top to
bottom layer, the sieves holes are from big to small. There are vibration
motors on both sides of the sieve trunk. While grain cleaning machine started, the sieving trunk begin vibration
sifter. Seeds falling on first layer then go through several sieve layers.
First layer sieve will remove the big impurity, final sieve layer sieve will
remove the small impurity. Middle layers can be used to remove either big,
small impurity or grading the cleaned seeds by size difference. Different layer
material has different outlets. Finally clients get cleaned and graded grain
seeds.
Model
Capacity (T/H)
Power (KW)
Weight (KG)
Feature
5XZC-3A
3
7.43
1300
Installed maize thresher
5XZC-3B
3
4.25
1200
5XZC-3C
3
7.25
1300
Installed wheat huller
5XZC-5A
5
12.74
1600
Installed maize thresher
5XZC-5CDH
5
11.74
1700
Installed wheat huller
5XZC-5DH
5
7.74
1600
With cyclone
5XZF-7.5F
7.5
10.1
1800
Double air cleaning systems,
with destoner plate
5XZC-15
10
10.5
1800
With cyclone
5XZC-15A
10
20.93
2200
Installed maize thresher
5XZC-25
20
19.45
2900
Double cyclones
5XZD-15AC
20
20.9
3000
Double sieve trunks

Seed Cleaner Cum Grader,Seed Cleaner Grader,Soybean Seed Cleaner,Air Screen Cleaner
SHIJIAZHUANG SYNMEC INTERNATIONAL TRADING LIMITED , https://www.seedgraincleaner.com